June 26, 2020
- Started again on nb-ros, but conda is not known. On my Shuttle, I could do conda activate pyhum.
- Yet, on Shuttle I don't have ~/git/PyHum, although I have the Linux-partition of nb-dual. On my nb-ros I have ~/git/PyHum. PyHum is installed, so I could do python -c "import PyHum;PyHum.dotest()" after export DISPLAY=127.0.0.1:0. The file ~/pyhum_test/test.DAT is read. No crash, only two warnings (plot could not be produced) for the last task:
===================================================
Input file is /home/arnoud/pyhum_test/test.DAT
Sonar file path is /home/arnoud/pyhum_test
cs2cs arguments are epsg:26949
Beam is 20.0 deg
pH is 7.0
Temperature is 10.0
Dwnward sonar freq. is 200.0
number of records for integration is 5
number of returned acoustic clusters is 3
drawing and printing map ...
drawing and printing map ...
plot could not be produced
drawing and printing map ...
plot could not be produced
- Assigned the letter U to the nb-dual partition, but this disk is only visible from Windows. Copied ~/git/PyHum from nb-dual via /mnt/c/tmp and to my Shuttle ~/git/PyHum.
- On nb-dual there were two version of my_test.py, one in ~/pyhum_test and one in my home-directory ~. Copied both to same location on my Shuttle. In ~ I am running the same experiment as my last call on May 8, 2020: python -c "import my_test; my_test.dotest(). Still same two warnings.
- Started the Gui with python -c "import PyHum;PyHum.gui()". Copied the ~/projects/MappingNature from nb-dual to Shuttle, including the MappingNature/data directory. Selected data/track1 as 998 model. The file is read without problems, although no additional print-statements. Selected data/track9 as Mega model. That fails on metadat = data.getmetadata() of PyHum/pyread.pyx
- Looked at Shuttle in ~/miniconda3/envs/pyhum/lib/python2.7/site-packages/PyHum. Most files are from May 8th, only the _gui.pyc is from May 18th.
- Also looked at nb-dual in ~/anaconda2/lib/python2.7/site-packages. There I have May 11 versions of _pyhum_e1e2.py and _pyhum_read.py. Copied those to Shuttle's miniconda3 environment. The result: _pyhum_read at least handles track9 gracefully:
Checking the epsg code you have chosen for compatibility with Basemap ...
... epsg code compatible
WARNING: Because files have to be read in byte by byte,
this could take a very long time ...
something went wrong with the parallelised version of pyread ...
something went wrong with getmetadate in old pyread ...
no number of datapoints metadata ...
no heading metadata ...
no n/e metadata ...
no bearing metadata ...
no dist_m metadata ...
no dep_m metadata ...
portside scan not available
starboardside scan not available
low-freq. scan not available
high-freq. scan not available
skipping several metadata keys
Processing took 27.2426819801seconds to analyse
Done!
May 24, 2020
- Reproducing the installation process of May 8th on nb-ros (Ubuntu 14.04), so that I can work this week on the code.
- First step: conda create --name pyhum python=2;conda activate pyhum.
- Second step: conda install -c conda-forge basemap-data-hires -y.
- Third step: conda install -c conda-forge gdal.
- Fourth step: conda install -c conda-forge scipy numpy scikit-image
- Fifth step: pip install simplekml sklearn pandas dask
- Sixth step: pip install joblib toolz cython
- Fifth step: pip install pyresample
- Last step: pip install PyHum --no-deps, installed successfully PyHum-1.4.6.
- Copied the project and github from nb-dual.
- Yet python -c "import PyHum;PyHum.dotest()" fails on scipy/linalg, which has undefined symbol PyFPE_buf. scipy is version 0.18.1. Yet, the version installed with conda is 1.2.1. A simple pip uninstall scipy solved this issue, the PyHum.dotest() works, yet with a warning that basemaps could not be imported!
- Same is true for PyHum.gui(), which also indicates that the epsg code of track1 is not compatible with Basemap.
- Checked the version with conda list, basemap is v1.3.0, while basemap-data-hires is v1.2.1. On my Shuttle I was using basemap-1.2.0.
- Trying to solve this issue with: conda install -c conda-forge basemap=1.2.1. It goes already wrong with import PyHum, which makes calls to from mpl_toolkits.basemap import Basemap. That import fails on numpy datasource. Now matplotlib fails on missing numpy. matplotlib 2.2.5 is installed via pip and matplotlib-base 2.2.5 is installed via conda. Performed both pip uninstall numpy and pip uninstall matplotlib, followed by conda install -c conda-forge matplotlib. That succeeds, but I don't see matplotlib in the side-packages.
- Could install simplekml also via conda -c conda-forge (v1.3.5). Tried pip install numpy scipy matplotlib --no-cache-dir, as suggested in this post. I receive numpy 1.16.6 and matplotlib 2.2.5
- The pip-install complains that pyhum-1.4.6 has requirement pyresample==1.1.4, while I have pyresample 1.14.0.
- PyHum.dotest runs again, with warning about Basemap.
- Trying conda install -c conda-forge basemap=1.2.0. Also gdal and geos will be downgraded, but no changes in numpy and/or matplotlib.
- According to conda, I have now numpy installed both via conda (1.16.5) and pip (1.16.6). Also matplotlib is v2.2.5, while on nb-dual it was v2.2.4. Install fails because no space left on device.
- I still had a very old version of numpy (1.8) installed on my system (outside the pyhum environment).
- Numpy is not installed in /usr/local/lib/python2.7/site-packages, but in ../dis-packages. The conda location is in ~/miniconda/pkgs
- Installed all packages that were requested via pip. Back to warning again. Installed basemap (1.2.1) without channel specifation. Finaly the Basemap warning is gone, although now the np.shape error is back.
May 11, 2020
- Reading track1 data as type 998-model: get expected output:
Read the son-file without idx
('Read %d sonar data packets of length %d', 4133, 2451)
Reading the son-files with idx
Read the son-file without idx
('Read %d sonar data packets of length %d', 4133, 2451)
Reading the son-files with idx
Read the son-file without idx
('Read %d sonar data packets of length %d', 2069, 2451)
Reading the son-files with idx
Read the son-file without idx
('Read %d sonar data packets of length %d', 2064, 2451)
('getsonar found: ', 'sidescan_port')
('data_port has length %d', 8266)
('packet has value %d', 2451)
('getsonar found: ', 'sidescan_port')
port sonar data will be parsed into 6.0, 99 m chunks
('getsonar found: ', 'sidescan_starboard')
starboard sonar data will be parsed into 6.0, 99 m chunks
('getsonar found: ', 'down_lowfreq')
low-freq. sonar data will be parsed into 6.0, 99 m chunks
('getsonar found: ', 'down_highfreq')
high-freq. sonar data will be parsed into 6.0, 99 m chunks
Processing took 97.6821660995seconds to analyse
When doing the same with track9, it crashes (probably due to huge packet value):
Reading the son-files with idx
('Read %d sonar data packets of length %d', 7424, 1255)
Reading the son-files with idx
('Read %d sonar data packets of length %d', 7424, 1255)
Reading the son-files with idx
('Read %d sonar data packets of length %d', 7424, 1255)
Reading the son-files with idx
('Read %d sonar data packets of length %d', 7425, 1255)
('getsonar found: ', 'unknown')
('data_port has length %d', 14848)
('packet has value %d', 721747968)
Segmentation fault (core dumped)
- The huge value is the value of k[0][12], so could overwrite this with the actual value 1255 (where in metadata is this value?).
- Copied the code of May 6 into the github directory, and put a check on k[0][13], but still something went wrong (although this was now again gracefully handled by _pyhum_read.py):
('getsonar found: ', 'unknown')
('data_port has length %d', 14848)
('packet has value %d', 721747968)
something went wrong with getmetadate in old pyread ...
no number of datapoints metadata ...
no heading metadata ...
no n/e metadata ...
no bearing metadata ...
no dist_m metadata ...
no dep_m metadata ...
('getsonar found: ', 'unknown')
portside scan not available
('getsonar found: ', 'unknown')
starboardside scan not available
('getsonar found: ', 'unknown')
low-freq. scan not available
('getsonar found: ', 'unknown')
high-freq. scan not available
skipping several metadata keys
- Strange enough I don't see my code changes anymore when running the script again, although installed in python2.7/site-packages.
May 8, 2020
- Tried to reproduce Lisa installation problems on my Ubuntu environment on my Shuttle.
- No problems with conda create --name pyhum python=2;source activate pyhum.
- Changed the order a bit, and first installed basemap with conda install -c conda-forge basemap-data-hires -y.
- Next conda install -c conda-forge gdal, which makes the following changes to the previous installation:
The following packages will be SUPERSEDED by a higher-priority channel:
basemap conda-forge::basemap-1.2.0-py27h673bf~ --> pkgs/main::basemap-1.2.0-py27h705c2d8_0
geos conda-forge::geos-3.7.1-hf484d3e_1000 --> pkgs/main::geos-3.6.2-heeff764_2
The following packages will be DOWNGRADED:
proj4 5.2.0-he1b5a44_1006 --> 4.9.3-h516909a_9
pyproj 1.9.6-py27h516909a_1002 --> 1.9.5.1-py27h2944ce7_1006
- No problems so far, continue with conda install scipy numpy scikit-image, which makes the following changes to the previous installation:
The following packages will be UPDATED:
numpy conda-forge::numpy-1.16.5-py27h95a140~ --> pkgs/main::numpy-1.16.6-py27h30dfecb_0
The following packages will be SUPERSEDED by a higher-priority channel:
ca-certificates conda-forge::ca-certificates-2020.4.5~ --> pkgs/main::ca-certificates-2020.1.1-0
certifi conda-forge::certifi-2019.11.28-py27h~ --> pkgs/main::certifi-2019.11.28-py27_0
openssl conda-forge::openssl-1.0.2u-h516909a_0 --> pkgs/main::openssl-1.0.2u-h7b6447c_0
- No problems, continue with pip install simplekml sklearn pandas dask, followed by pip install joblib toolz cython and pip install pyresample.
- Also pip install PyHum --no-deps works without problems.
- Even python -c "import PyHum;PyHum.dotest()" seems to work:
Input file is ~/pyhum_test/test.DAT
Son files are in ~/pyhum_test
cs2cs arguments are epsg:26949
Draft: 0.3
Celerity of sound: 1450.0 m/s
Port and starboard will be flipped
Transducer length is 0.108 m
Bed picking is auto
Only 1 chunk will be produced
Data is from the 998 series
Bearing will be calculated from coordinates
Bearing will be filtered
Checking the epsg code you have chosen for compatibility with Basemap ...
... epsg code compatible
WARNING: Because files have to be read in byte by byte,
this could take a very long time ...
Processing took 17.299973011seconds to analyse
Done!
===================================================
Input file is ~/pyhum_test/test.DAT
Sonar file path is ~/pyhum_test
Max. transducer power is 1000.0 W
pH is 7.0
Temperature is 10.0
Processing took 18.5526208878seconds to analyse
Done!
===================================================
Input file is ~/pyhum_test/test.DAT
Sonar file path is ~/pyhum_test
Window is 31 square pixels
Threshold dissimilarity (shadow is <) is 3
Threshold correlation (shadow is <) is 0
Threshold contrast (shadow is <) is 6
Threshold energy (shadow is >) is 0
Threshold mean intensity (shadow is <) is 4
Processing took 30.3947219849seconds to analyse
Done!
===================================================
Input file is ~/pyhum_test/test.DAT
Sonar file path is ~/pyhum_test
Window is 10 square pixels
Number of sediment classes: 8
[Default] Number of processors is 6
processing port side ...
processing starboard side ...
Plotting ...
Processing took 73.3709928989seconds to analyse
Done!
===================================================
Input file is ~/pyhum_test/test.DAT
Sonar file path is ~/pyhum_test
cs2cs arguments are epsg:26949
Gridding resolution: 0.05
Mode for gridding: 1
Number of nearest neighbours for gridding: 64
Threshold number of standard deviations in sidescan intensity per grid cell up to which to accept: 5
Number of chunks for mapping: 1479
getting point cloud ...
writing point cloud
gridding ...
error: geotiff could not be created... check your gdal/ogr install
creating kmz file ...
drawing and printing map ...
Processing took 381.660418987seconds to analyse
Done!
===================================================
Input file is /home/arnoud/pyhum_test/test.DAT
Sonar file path is /home/arnoud/pyhum_test
cs2cs arguments are epsg:26949
Beam is 20.0 deg
pH is 7.0
Temperature is 10.0
Dwnward sonar freq. is 200.0
number of records for integration is 5
number of returned acoustic clusters is 3
drawing and printing map ...
drawing and printing map ...
plot could not be produced
drawing and printing map ...
plot could not be produced
- Everything seems to go right, except the geotiff and plot error (should try to install geotiff).
- Also PyHum.gui() runs without problems.
- Installed geotiff with conda install -c conda-forge geotiff, which makes the following changes to the previous installation:
The following NEW packages will be INSTALLED:
geotiff conda-forge/linux-64::geotiff-1.4.2-hfe6da40_1005
The following packages will be UPDATED:
ca-certificates pkgs/main::ca-certificates-2020.1.1-0 --> conda-forge::ca-certificates-2020.4.5.1-hecc5488_0
certifi pkgs/main::certifi-2019.11.28-py27_0 --> conda-forge::certifi-2019.11.28-py27h8c360ce_1
The following packages will be SUPERSEDED by a higher-priority channel:
openssl pkgs/main::openssl-1.0.2u-h7b6447c_0 --> conda-forge::openssl-1.0.2u-h516909a_0
- Installing geotiff didn't help, still same errors/warnings. Tried in python from osgeo import gdal, but that fails.
- Problem seems to be the loading of the poppler library, a bug that is solved in gdal 2.2.3. Unfortunatelly my version of poppler is 0.67 and gdal 2.2.2.
- Tried to install conda install -c esri gdal=2.3.3 but many conflicts where found.
- Tried instead to downgrade poppler with conda install -c conda-forge poppler=0.52.0. Also here many conflicts.
- Added the channels esri and conda-forge. Did a conda search -f gdal. Saw that gdal v2.2.4 was available in main. Did conda install gdal=2.2.4. As a result:
The following packages will be downloaded:
package | build
---------------------------|-----------------
gdal-2.2.4 | py27h637b7d7_1 973 KB
json-c-0.13.1 | h14c3975_1001 71 KB conda-forge
libgdal-2.2.4 | hc8d23f9_1 10.0 MB
libpq-10.6 | h13b8bad_1000 2.5 MB conda-forge
python-2.7.15 | h938d71a_1004 11.9 MB conda-forge
------------------------------------------------------------
Total: 25.4 MB
Still cannot import gdal due to poppler.
- Tried conda install gdal=2.3.3. As a result:
The following packages will be downloaded:
package | build
---------------------------|-----------------
basemap-1.2.0 | py27hf62cb97_3 15.2 MB conda-forge
cairo-1.14.12 | he6fea26_5 1.3 MB conda-forge
curl-7.68.0 | hf8cf82a_0 137 KB conda-forge
fontconfig-2.13.0 | h9420a91_0 227 KB
gdal-2.3.3 | py27hbb2a789_0 994 KB
geotiff-1.4.3 | hb6868eb_1001 1.1 MB conda-forge
hdf5-1.10.4 |nompi_h3c11f04_1106 5.3 MB conda-forge
kealib-1.4.7 | hd0c454d_6 154 KB
krb5-1.16.4 | h2fd8d38_0 1.4 MB conda-forge
libcurl-7.68.0 | hda55be3_0 564 KB conda-forge
libdap4-3.19.1 | h6ec2957_0 1.0 MB
libgdal-2.3.3 | h2e7e64b_0 11.1 MB
libnetcdf-4.6.2 | hbdf4f91_1001 1.3 MB conda-forge
libpq-11.4 | h4e4e079_0 2.5 MB conda-forge
libspatialite-4.3.0a | hb5ec416_1026 3.1 MB conda-forge
libssh2-1.8.2 | h22169c7_2 257 KB conda-forge
libuuid-1.0.3 | h1bed415_2 15 KB
matplotlib-base-2.2.4 | py27hfd891ef_0 6.6 MB conda-forge
openssl-1.1.1g | h516909a_0 2.1 MB conda-forge
pillow-6.2.1 | py27hd70f55b_1 625 KB conda-forge
poppler-0.67.0 | h4d7e492_3 8.5 MB conda-forge
python-2.7.15 | h721da81_1008 12.8 MB conda-forge
scikit-image-0.14.3 | py27hb3f55d8_0 24.0 MB conda-forge
xorg-libsm-1.2.2 | h470a237_5 24 KB conda-forge
------------------------------------------------------------
Total: 100.3 MB
Still from osgeo.gdal import * fails on poppler.so
- Trying conda install gdal=2.4.4, but get many conflicts.
- Instead, try to conda install poppler-data=0.4.7 and poppler-data=0.4.8, still no import.
- Trying conda install poppler=0.69.0. Still no import
- Tried to something completely different with sudo apt-get install -y poppler-utils. This is installs libpoppler73. Yet, the import complains on libpoppler.so.76.
- Tried conda install poppler=0.76.0, but this fails on many conflicts (based on python3).
- I found that libpoppler.so.76 is part of conda package v0.65. The different libpoppler.so versions installed by conda can be found with ls ~/miniconda3/pkg/poppler-*/lib. I had libraries so.78 and so.80.
- Tried conda install poppler=0.65.0, but this also leads to conflicts.
- Deactivate pyhum environment and created a new environment poppler. Now libpoppler.so.76 is in ~/miniconda3/pkgs
- Still, this library cannot be found, even when I copied the so.76 to ~/miniconda3/envs/pyhum/lib/python2.7/side-packages and/or side-packages/osgeo didn't help, but copying it to ~/miniconda3/envs/pyhum/lib/ solved my issue!
- Got an error:
File "/home/arnoud/miniconda3/envs/pyhum/lib/python2.7/lib-tk/Tkinter.py", line 1819, in __init__
self.tk = _tkinter.create(screenName, baseName, className, interactive, wantobjects, useTk, sync, use)
_tkinter.TclError: no display name and no $DISPLAY environment variable
but that was because in this environment I had not set export DISPLAY=127.0.0.1:0. When the DISPLAY was said the processing of PyHum.test() continues.
- No gdal/ogr error anymore during the gridding PyHum.test(), but still a warning in the last test: no plot can be produced:
gridding ...
creating kmz file ...
drawing and printing map ...
Processing took 419.456223965seconds to analyse
Done!
===================================================
Input file is /home/arnoud/pyhum_test/test.DAT
Sonar file path is /home/arnoud/pyhum_test
cs2cs arguments are epsg:26949
Beam is 20.0 deg
pH is 7.0
Temperature is 10.0
Dwnward sonar freq. is 200.0
number of records for integration is 5
number of returned acoustic clusters is 3
drawing and printing map ...
drawing and printing map ...
plot could not be produced
drawing and printing map ...
plot could not be produced
- The last warning is printed in the function e1e2, with a plt.figure and custom_save are called.
- Tried to reproduce the error on nb-dual, but PyHum.correct still fails on shape mismatch. Strange, both have numpy v1.16.6. The two environments are not completely the same. At nb-dual cython is installed via conda, instead of pip. geos is v3.6.2 instead of v3.7.1 on the Shuttle. scikit-image is 0.14.3 on the Shuttle, while 0.14.5 on nb-dual.
- Made a version with only e1e2() in ~/my_test.py and calling it with python -c "import my_test; my_test.dotest(). There are 6 places where drawing is called. In drawing #3 it goes well, in #4 and #5 no plot could be produced. In both cases it goes wrong with the custom_save. custom_save() is just plt.savefig.
- When adding a plt.show() before drawing #4 six figures are drawn. In the 6th figure a type-error is made (casting dtype('S1') to dtype('dfloat64')). Removed the scatter in drawing #4, no type-error anymore (but empty png). Strange, because same scatter-call is made in drawing #3b, the only difference is rough.flatten vs hard.flatten
May 6, 2020
- Printed k[0][13] in getsonar function, and in all cases (of track9) the string is 'unknown'.
- Read the information around k[0][13]:
('getsonar k[0][12]: ', 721747968)
('getsonar k[0][13]: ', 'unknown')
('getsonar k[0][14]: ', 'Coroutine')
('getsonar k[1][13]: ', 178)
- Also printed the other fields:
('getsonar k[0][0]: ', 1)
('getsonar k[0][1]: ', 1)
('getsonar k[0][2]: ', 566206)
('getsonar k[0][3]: ', 6847765)
('getsonar k[0][4]: ', 2)
('getsonar k[0][5]: ', 18.0)
('getsonar k[0][6]: ', 1)
('getsonar k[0][7]: ', 0.8)
('getsonar k[0][8]: ', 1.9)
('getsonar k[0][9]: ', 161)
('getsonar k[0][10]: ', 195)
('getsonar k[0][11]: ', -1985891.645)
('getsonar k[0][12]: ', 721747968)
('getsonar k[0][13]: ', 'unknown')
('getsonar k[0][14]: ', 269)
('getsonar k[0][15]: ', 2797)
Core dump on k[0][16]
- Trying to interpret this fields. It is not the 12 fields which is read from .DAT file by data.gethumdat()
- It is also not the 9 fields as read by data.getmetadata(), although the floats at field 5, 7, 8 could be interpreted as values for depth and spd.
- You would expect that the first 22 bytes of SON files are read. Byte 42 should be an integer which contains the beam-number (2 is SI Port, 3 is SI Starboard).
- Checked both the .DAT and .SON files with strings, but no visible 'sidescan' or 'unknown' are visible.
May 4, 2020
- Activated conda activate pyhum. The command python -c "import PyHum" works. Yet, python pyhum_test/my_test.py fails on FileOpeners().
- Started my script ~/bin/pyhum.sh to call the gui, which works fine on the ~/pyhum_test/test.DAT. Only the Done! once read is a bit unneccessary.
- The gui crashes on track9 on the pyread.pyx line 532 an 538.
- Removed the call to metadata on line 426 of _pyhum_read.py, but than it fails on line 431 (no metadat object).
- The metadata is created in the C-file ~/git/PyHum/PyHum/pyread.pyx. The datastructure is dictionary with fields (lat, lon, spd, time_s, e, n, dep_m, calltime, heading). The call of getmetadata fails on data_port = self._getsonar('sidescan_port').
- Added a try around getsonar, build the c-file with cython pyread.pyx and copied that to ~/anaconda2/envs/pyhum/lib/python2.7/site-packages/PyHum with the command python setup.py install.
- The _pyhum_read.py in the site-package was overwritten from the github code, but the _pyhum_read.pyc was still used (skipping the metadata read). Called the gui script with from conda pyhum environment, and the new code is called. Do not see the print, so getsonar returned succesfully, but print length failed.
- Next call to fail will probably tmp = np.squeeze(data_port). squeeze removes all singleton dimensions, converting the (1, n) vector to an n-length array.
- Tried to read the track1 code, to see the data_port length, but my code is not called (I am in an exception branch).
- For track9 the code is called. Nicely the exception couldn't read data_port is printed, followed by an crash on line 566 (index 2 out of bounds).
- Added a getsonar to the parallel version, now track1 gives the output:
something went wrong with the parallelised version of pyread ...
('data_port has length %d', 8266)
('packet has value %d', 2451)
- For the test.dat the output is:
('data_port has length %d', 6904)
('packet has value %d', 1479)
- Put several protections in py_read.py. Now get warnings:
no number of datapoints metadata ...
no heading metadata ...
no n/e metadata ...
no bearing metadata ...
- Now fails on 481 (variable lat not assigned).
- Looked into getsonar(). Added print adding integers, which each time has length 2451 (packet length) for track1. Is called many times (8266x ?)
- No getsonar print for track9, protection is working for the next calls (until shape_port on line 683).
- track 1 still successfull read:
Reading the son-files with idx
Read the son-file without idx
('Read %d sonar data packets of length %d', 4133, 2451)
('Read %d sonar data packets of length %d', 4133, 2451)
('Read %d sonar data packets of length %d', 2069, 2451)
('Read %d sonar data packets of length %d', 2064, 2451)
('data_port has length %d', 8266)
('packet has value %d', 2451)
port sonar data will be parsed into 6.0, 99 m chunks
starboard sonar data will be parsed into 6.0, 99 m chunks
low-freq. sonar data will be parsed into 6.0, 99 m chunks
high-freq. sonar data will be parsed into 6.0, 99 m chunks
Seems that data_port is the data packets of port- and starboard-side combined.
This should be compared with Track1 has 2069 records.
Each record has 2518 bytes. That is a difference of 67 bytes. Corresponds with headbytes of model 997.
- The son-files are also read for track9, although not recognized as sonardata:
Reading the son-files with idx
('Read %d sonar data packets of length %d', 7424, 1255)
('Read %d sonar data packets of length %d', 7424, 1255)
('Read %d sonar data packets of length %d', 7424, 1255)
('Read %d sonar data packets of length %d', 7425, 1255)
Couldn't read data_port in getmetadata
Earlier I saw that track9 has resp. 7430, 7429, 7429 and 7429 records. Each record is 1323 bytes. That is a difference (?dataheader of 68 bytes).
- Initialized metadat as empty dictonairy, instead of empty list. Fails in line 463, which means that the checks before that line worked. Added some extra protection, the metadata function finishes gracefully:
Couldn't read data_port in getmetadata
something went wrong with getmetadate in old pyread ...
no number of datapoints metadata ...
no heading metadata ...
no n/e metadata ...
no bearing metadata ...
no dist_m metadata ...
no dep_m metadata ...
portside scan not available
starboardside scan not available
low-freq. scan not available
high-freq. scan not available
skipping several metadata keys
Processing took 28.433989048seconds to analyse
Done!
===================================================
Done! Read module finished
- A Rec00009rawdat.csv is created, but only contains a header of the colums, no rows of data is included.
April 22, 2020
- Loaded the kml file of the pyhum_test in My Maps. The sonar was recorded at Horeshoe Bend in the Colorado river.
- The kml file from the track1 was recorded for the coast of the Miami Sea acquarium.
- The kml file of track9 is not generated yet.
April 21, 2020
- Looked at the error which occurs for reading track9. It goes wrong in line 532, getmetadata. In this function getsonar('sidescan_port') is called, with a fixed packet data_port[0][12]. The getmetadata is called in pyhum_read.py, at line 426 (non-parallel branch). The TypeError seems to be on self or data_port.
- The print "Done!" is the last statement (in a tkMessageBox) of the function _proc (called by hitting the button processing).
- Replaced the tkMessageBox statement with a print statement, still the Gui exits (gracefully) after processing. Further nothing extra than the frames and the mainloop. Removed the self.update()
- With the parameter settings of test.py, I was able to perform a PyHum.read() on test.DAT.
- Tried the same on track1 (still 998). Now something went wrong with the parallelised version of pyread. The result are several new *.png in the SON directory. Also several png from yesterday are still there!
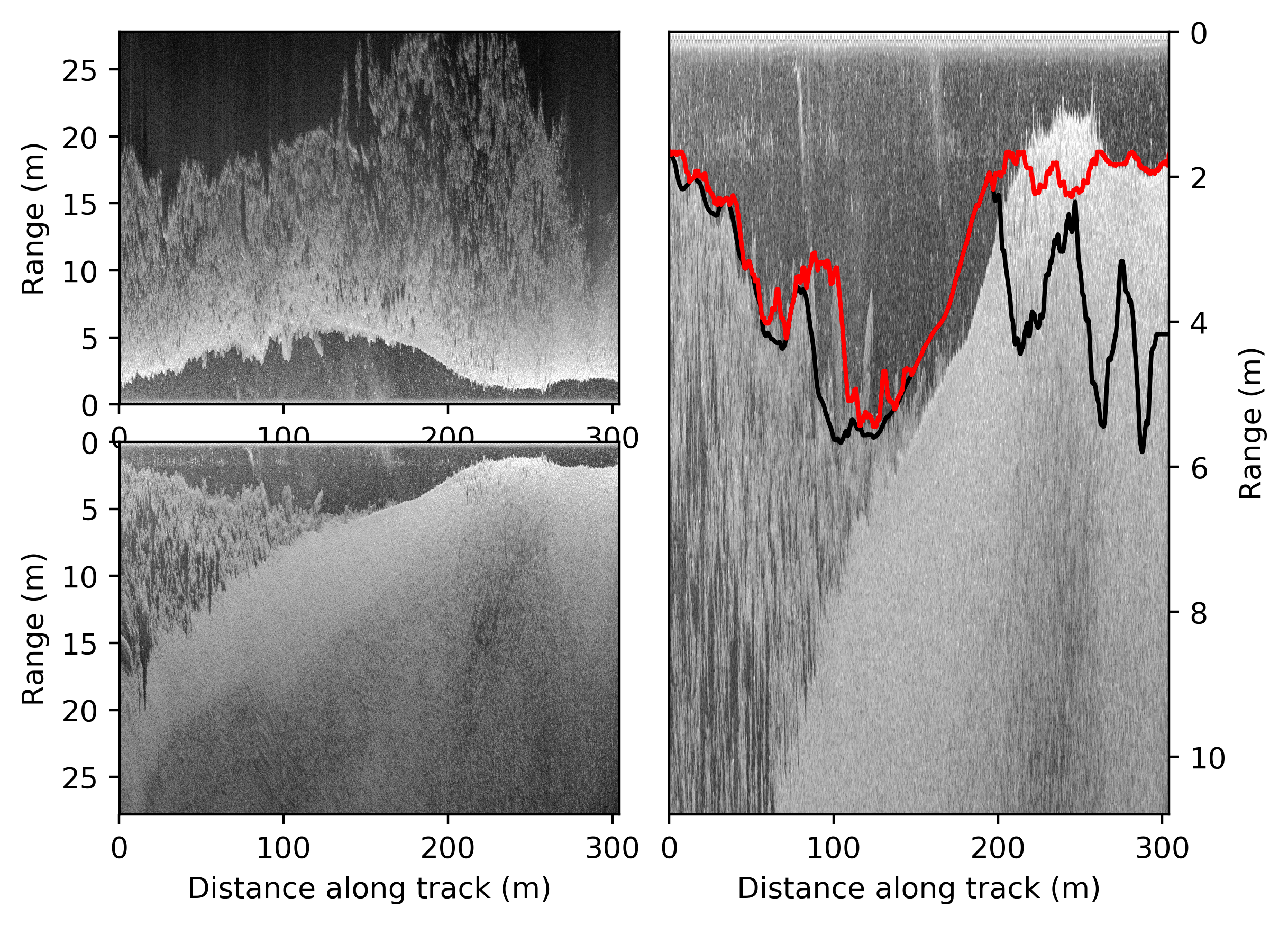
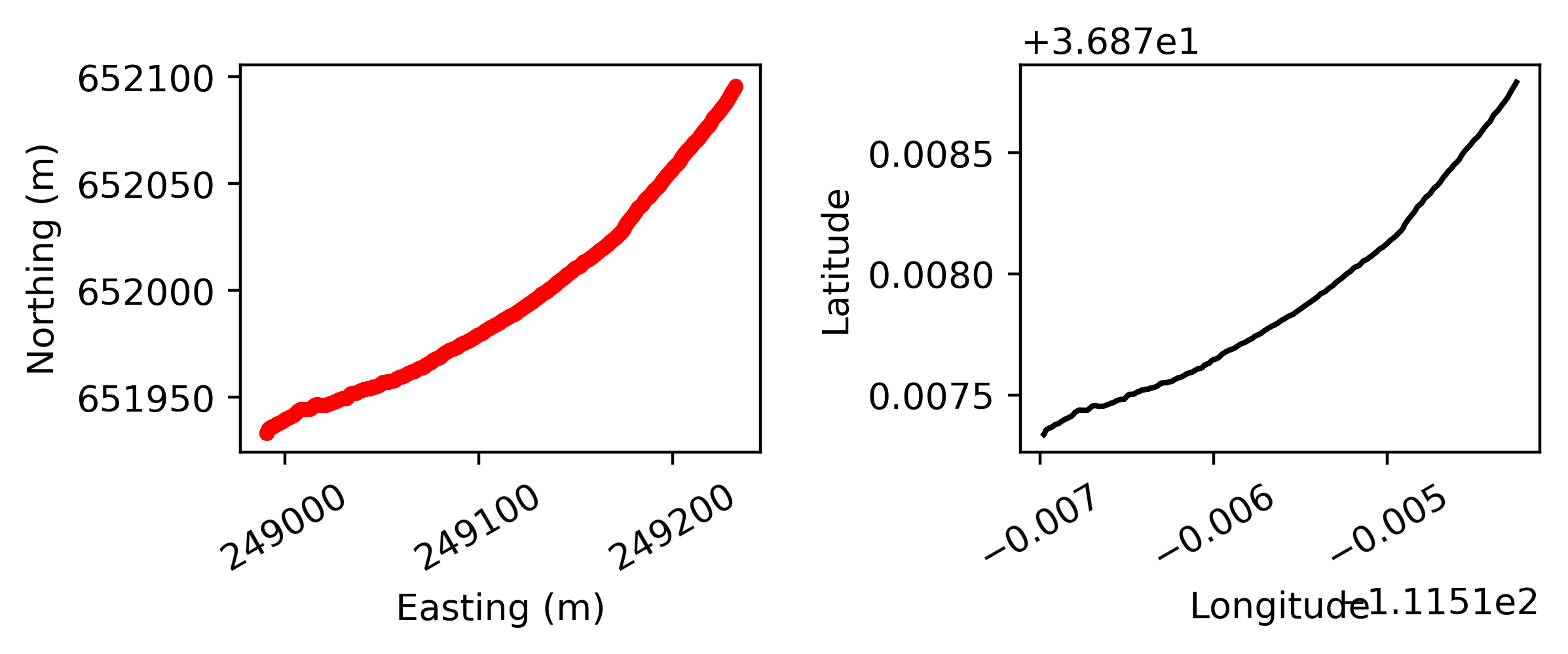
- The results of pyhum_test data look like:
- The results of track1 data look like:
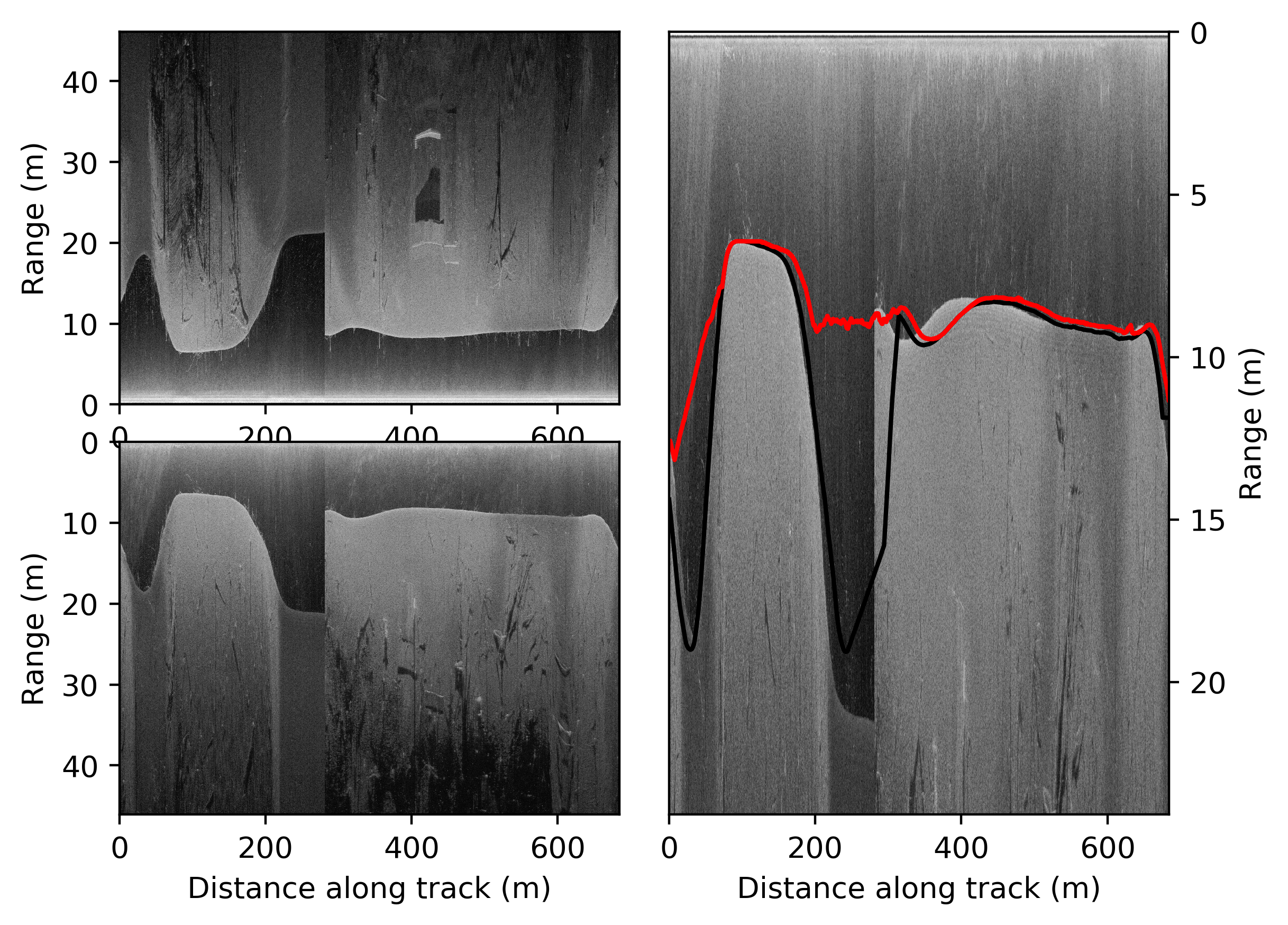
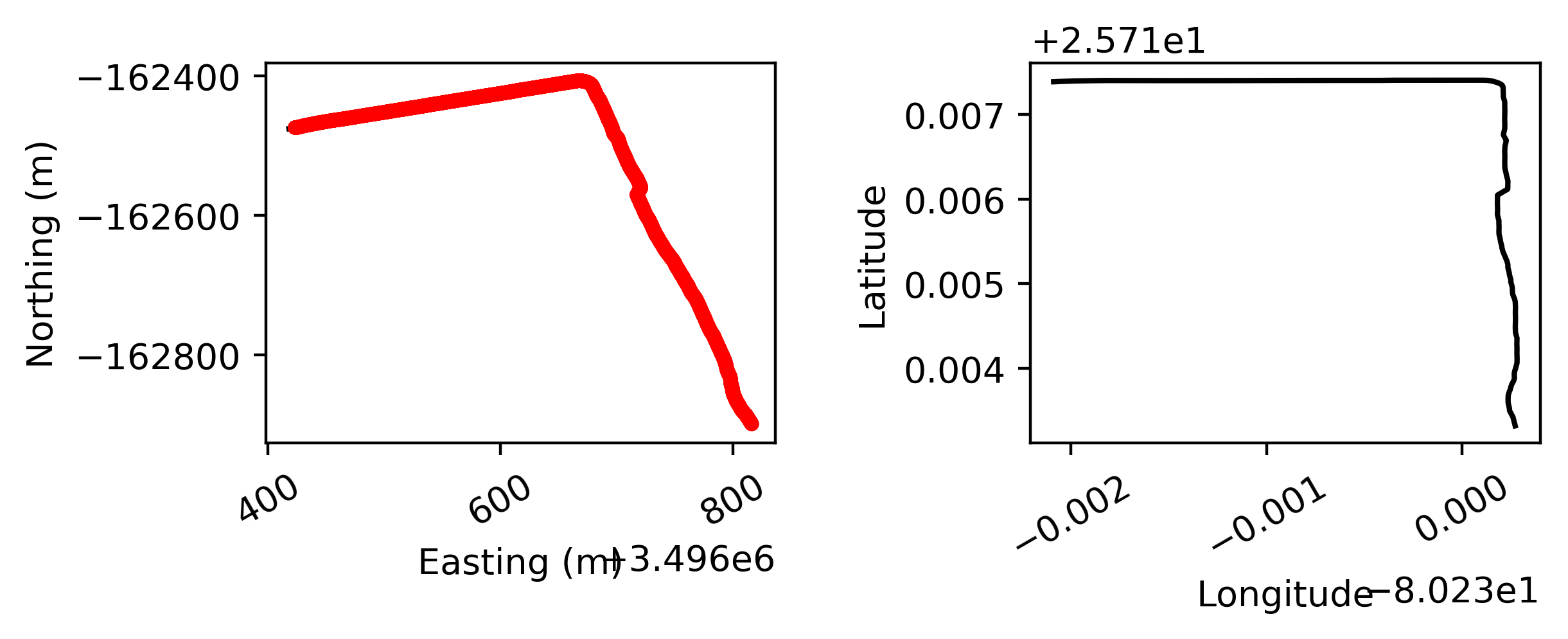
At the left you see the two side sonars, next to the sonar directly under the boat. On the right the trajectory that is followed according to the gps is given.
- Tried also track9 with model=2. Same error in reading getmetadata.
April 20, 2020
- Followed the instructions from PyHum
- Downloaded the two datasets (track1 and track9). Only 359 Mb disk space left.
- A lot of conflicts for conda install -c conda-forge basemap-data-hires -y. There are only 4 versions available, the version of the channel conda-forge is v1.2.1, on channel anaconda is v1.2.0 available.
- For basemap only Windows installation files are available, for Linux one should install from source.
- Removed UnrealEngine 4.24 to get some space on my Linux partition of nb-dual.
- Installation of basemap-data-hires without specifying a channel works (yet it is v1.2.0).
- Continued with the installation of PyHum from source with command cd ~/git/PyHum; python setup.py install. That works, the package is installed in ~/anaconda2/lib/python2.7/site-packages (PyHum-1.4.6).
- Also cloned basemap into ~/git, but a build was not needed after the successful conda install basemap-data-hires.
- The simple test python -c "import PyHum; PyHum.test.dotest()", but that was because in my second terminal I hadn't activated my conda environment. I installed the other packages in my conda base environment, so had to add them also to pyhum environment. Yet, there the conflicts are back!
- Removed gdal, that works, I can install basemap. After pip install cython I can do python setup.py install in ~/git/PyHum. Continued with python -c "import PyHum; PyHum.test.dotest()". Added with pip install missing packages like scipy, simplekml, sklearn and joblib. Still, problems with import ppdrc. Problem seems that I was still in the source directory. Outside that the directory the installed PyHum packages are used.
- Had to pip install the packages scikit-image, pyresample and pandas. Test put a test sonar file in ~/pyhum_test, processed it until Temperature is 10.0, but failed on line 138 of test.py (in function remove_water np.expand_dims() received a shape (1479,1,1) instead of (1,1479).
- The Gui command python -c "import PyHum; PyHum.gui()" works, but I should first downgrade my 4K resolution.
- Was able to go to tab Read and specified the DAT from track1. Now I have to select the Humminbird model! Selected as first choice the 997 series as model. That seems to work, although the process terminates when the reading is done:
Data is from the 997 series
Checking the epsg code you have chosen for compatibility with Basemap ...
... epsg code compatible
WARNING: Because files have to be read in byte by byte,
this could take a very long time ...
port sonar data will be parsed into 6.0, 99 m chunks
starboard sonar data will be parsed into 6.0, 99 m chunks
low-freq. sonar data will be parsed into 6.0, 99 m chunks
high-freq. sonar data will be parsed into 6.0, 99 m chunks
Processing took 71.3536579609seconds to analyse
Done!
- Track 9 is from a Humminbird Solix 12 CHIRP Mega SI, track1 is probably from a 997CX2 (from HumViewer).
- Also crashes after selecting data in ~/pyhum_test
- Tried the track9 data with as model MEGA series. First part went well, second part gave an error:
cs2cs arguments: epsg:26949
chunk argument: d100
Input file is ~/projects/MappingNature/data/track9/Rec00009.DAT
Son files are in ~/projects/MappingNature/data/track9/Rec00009
cs2cs arguments are epsg:26949
Draft: 0.3
Celerity of sound: 1450.0 m/s
Transducer length is 0.108 m
Bed picking is auto
Chunks based on distance of 100 m
Data is from the MEGA series
Checking the epsg code you have chosen for compatibility with Basemap ...
... epsg code compatible
WARNING: Because files have to be read in byte by byte,
this could take a very long time ...
something went wrong with the parallelised version of pyread ...
- The error was in pyhum_read.py, line 426 (data.getmetadata()). In line 538 of getmetadata I recieve NoneType is not subscriptable. The GUI doesn't crash after the failed read!
- Removed the crashing line test.py. The test reads correctly (reports Humminbird model 998). Yet, the test of site-packages is called. When going into the directory with os.chdir("~/pyhum_test") got a lot of import orders, but still the function from site-packages is called. Calling import test; test.dotest() solves that. Now the next call fails (because there is no test_data_port_la.dat). There is test_data_port.dat in this directory, maybe it is created in PyHum.correct(). Yes, that is specified in PyHum reference manual.
- Also texture2 needs test_data_port_la.dat!
- Scanned the User manual of the Humminbird, the Solix can communicate with other sensors via the NMEA 0183 protocol (p. 244-245), such as the DBT - Depth Below Transducer and MTW - Water Temperature.
- The specification of the SON files is available on openseamap.org. The Header is known, the Data still has to be discovered!
- This is what Daniel Buscombe has discovered: How PyHum reads Humminbird files
- Installed hexedit and inspected ~/pyhum_test/test.DAT. Shows 4x16 bytes, terminated by a 0F.
- Looked into pyread.pyx. If the idx is corrupted, the whole file is interpreted as integers, read as big-endian unsigned chars (struct.unpack('>B)).
- With the Knuth-Morris-Pratt algorithm, the start sequence is found (192,222,171,33,128): in Hex this is the sequence(C0 DE AB 21 80). I see this pattern in ~/pyhum_test/B001.SON at position 0x60A.
- In track1/B000.SON I see this pattern at position 0x9D6.
- In track9/B001.SON I see this pattern at position 0x52B.
- In track9/B002.SON I see this pattern at position 0x52B.
- In track9/B003.SON I see this pattern at position 0x52B.
- In track9/B004.SON I see this pattern at position 0x52B.
- Cloned binwalk. With the command binwalk -R "\xc0\xde\xab\x21\x80" B001.SON | wc -l reveiled that there are 1731 entries, the first at position 0x60A.
- Track1 has 2069 records.
- Track9 has resp. 7430, 7429, 7429 and 7429 records. Each record is 1323 bytes.
- In track1 each record has 2518 bytes




