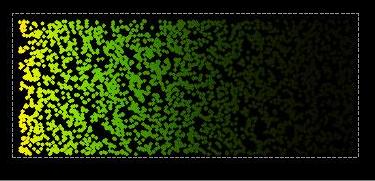
 Two-dimensional simulation of a bicoid gradient in Drosophila
Two-dimensional simulation of a bicoid gradient in Drosophila 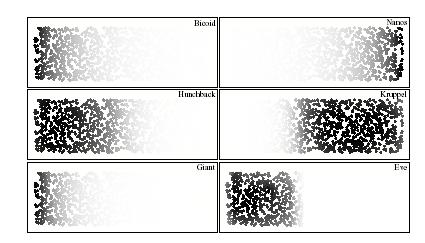 Two-dimensional simulation of gradients involved in the specification of Eve stripe two
in Drosophila
Two-dimensional simulation of gradients involved in the specification of Eve stripe two
in Drosophila Simulation of developmental regulatory networks
 Two-dimensional simulation of a bicoid gradient in Drosophila Two-dimensional simulation of a bicoid gradient in Drosophila |
 Two-dimensional simulation of gradients involved in the specification of Eve stripe two
in Drosophila Two-dimensional simulation of gradients involved in the specification of Eve stripe two
in Drosophila |
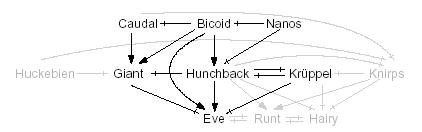 Regulatory network involved in the specification of Eve stripe two in Drosophila Regulatory network involved in the specification of Eve stripe two in Drosophila |
The project
The project is financed by the Netherlands Organisation for Scientific
Research, NWO, research program Computational Life Sciences and consists
of 2 Ph.D. positions, one (PhD modelling regulatory developmental networks) at the Section Computational Science
of the Universiteit van Amsterdam (UvA)
(http://www.science.uva.nl/research/scs/)
and one (PhD software development, numerical analysis) at the Center for Mathematics and Computer Science (CWI)
in Amsterdam
(http://www.cwi.nl/htbin/pdels/frame?RegNet)
and several other researchers from the UvA, CWI and two researchers from molecular developmental biology (see
below: ``The research team''). This project is carried out within the framework of the Silicon Cell
Initiative Amsterdam (http://www.siliconcell.net/sica/)
Abstract
Genetic regulation plays a fundamental role in biological processes.
Regulatory systems cannot simply be described as an assembly of
genes and proteins and diagrams of their interconnections. Many analysis
techniques, as for example cluster analysis, only provide `correlations'
between genes and do not provide insight into causal relations between
the genes in a regulatory network. An important option for the analysis
of regulatory control systems are simulation models in combination with
optimization algorithms.
In this project we will develop a model for simulating regulatory networks
that are capable of quantitatively reproducing spatial and temporal
expression patterns in developmental processes. The model is a
generalization of the standard connectionist model used for modelling
genetic interactions. The model will be coupled with a biomechanical
model of cell aggregates and used to study the formation of spatial
and temporal expression patterns of gene products during development in
cellular systems. As a case study we are planning to use the body plan formation
in relatively simple multi-cellular organisms (sponges and sceleractinian
corals).
Mathematically speaking this amounts to continuum-discrete hybrid models
where discrete, moving and deformable objects in which biochemical reactions
take place exchange species with the surrounding environment modelled as
a continuum in which species diffuse and decay.
A major issue are correct estimations of the parameter settings
in the network model (the regulatory weight factors). Therefore the
model will be used in combination with optimization algorithms
(genetic algorithms and simulated annealing) to explore large parameter
spaces of regulatory networks and to select specific
spatial and temporal expression patterns.
Collaboration with Russian researchers
We collaborate in this project with the research groups from Prof. Dr Alexander M. Samsonov, (Theoretical Department
The Ioffe Institute of the Russian Academy of Sciences, St.Petersburg, Russia) and
Prof Dr. Maria Samsonova (St. Petersburg State Polytechnical University St. Petersburg, Russia).
This collaboration is funded by the Netherlands Organisation for Scientific
Research and the Russian Science Foundation. The website of this collaboration is
located at:
http://http://urchin.spbcas.ru/sponges/
The Research Team
Section Computational Science (UvA)