Factor graph (.fg) file format
This section describes the .fg file format used in libDAI to store factor graphs. Markov Random Fields are special cases of factor graphs, as are Bayesian networks. A factor graph can be specified as follows: for each factor, one has to specify which variables occur in the factor, what their respective cardinalities (i.e., number of possible values) are, and a table listing all the values of that factor for all possible configurations of these variables.
A .fg file is not much more than that. It starts with a line containing the number of factors in that graph, followed by an empty line. Then all factors are specified, using one block for each factor, where the blocks are seperated by empty lines. Each variable occurring in the factor graph has a unique identifier, its label (which should be a nonnegative integer). Comment lines which start with # are ignored.
Factor block format
Each block describing a factor starts with a line containing the number of variables in that factor. The second line contains the labels of these variables, seperated by spaces (labels are nonnegative integers and to avoid confusion, it is suggested to start counting at 0). The third line contains the number of possible values of each of these variables, also seperated by spaces. Note that there is some redundancy here, since if a variable appears in more than one factor, the cardinality of that variable appears several times in the .fg file; obviously, these cardinalities should be consistent. The fourth line contains the number of nonzero entries in the factor table. The rest of the lines contain these nonzero entries; each line consists of a table index, followed by white-space, followed by the value corresponding to that table index. The most difficult part is getting the indexing right. The convention that is used is that the left-most variables cycle through their values the fastest (similar to MatLab indexing of multidimensional arrays).
Example
An example block describing one factor is:
3 4 8 7 3 2 2 11 0 0.1 1 3.5 2 2.8 3 6.3 4 8.4 6 7.4 7 2.4 8 8.9 9 1.3 10 1.6 11 2.6
which corresponds to the following factor:
![\[ \begin{array}{ccc|c} x_4 & x_8 & x_7 & \mbox{value}\\ \hline 0 & 0 & 0 & 0.1\\ 1 & 0 & 0 & 3.5\\ 2 & 0 & 0 & 2.8\\ 0 & 1 & 0 & 6.3\\ 1 & 1 & 0 & 8.4\\ 2 & 1 & 0 & 0.0\\ 0 & 0 & 1 & 7.4\\ 1 & 0 & 1 & 2.4\\ 2 & 0 & 1 & 8.9\\ 0 & 1 & 1 & 1.3\\ 1 & 1 & 1 & 1.6\\ 2 & 1 & 1 & 2.6 \end{array} \]](form_48.png)
Note that the value of 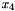 changes fastest, followed by that of
changes fastest, followed by that of 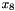 , and
, and 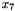 varies the slowest, corresponding to the second line of the block ("4 8 7"). Further,
varies the slowest, corresponding to the second line of the block ("4 8 7"). Further, 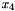 can take on three values, and
can take on three values, and 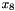 and
and 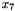 each have two possible values, as described in the third line of the block ("3 2 2"). The table contains 11 non-zero entries (all except for the fifth entry). Note that the eleventh and twelveth entries are interchanged.
each have two possible values, as described in the third line of the block ("3 2 2"). The table contains 11 non-zero entries (all except for the fifth entry). Note that the eleventh and twelveth entries are interchanged.
A final note: the internal representation in libDAI of the factor above is different, because the variables are ordered according to their indices (i.e., the ordering would be 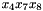 ) and the values of the table are stored accordingly, with the variable having the smallest index changing fastest:
) and the values of the table are stored accordingly, with the variable having the smallest index changing fastest:
![\[ \begin{array}{ccc|c} x_4 & x_7 & x_8 & \mbox{value}\\ \hline 0 & 0 & 0 & 0.1\\ 1 & 0 & 0 & 3.5\\ 2 & 0 & 0 & 2.8\\ 0 & 1 & 0 & 7.4\\ 1 & 1 & 0 & 2.4\\ 2 & 1 & 0 & 8.9\\ 0 & 0 & 1 & 6.3\\ 1 & 0 & 1 & 8.4\\ 2 & 0 & 1 & 0.0\\ 0 & 1 & 1 & 1.3\\ 1 & 1 & 1 & 1.6\\ 2 & 1 & 1 & 2.6 \end{array} \]](form_53.png)
Evidence (.tab) file format
This section describes the .tab fileformat used in libDAI to store "evidence", i.e., a data set consisting of multiple samples, where each sample is the observed joint state of some variables.
A .tab file is a tabular data file, consisting of a header line, followed by an empty line, followed by the data points, with one line for each data point. Each line (apart from the empty one) should have the same number of columns, where columns are separated by one tab character. Each column corresponds to a variable. The header line consists of the variable labels (corresponding to dai::Var::label()). The other lines are observed joint states of the variables, i.e., each line corresponds to a joint observation of the variables, and each column of a line contains the state of the variable associated with that column. Missing data is handled simply by having two consecutive tab characters, without any characters in between.
Example
1 3 2
0 0 1 1 0 1 1 1
This would correspond to a data set consisting of three observations concerning the variables with labels 1, 3 and 2; the first observation being 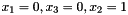 , the second observation being
, the second observation being 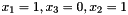 , and the third observation being
, and the third observation being 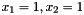 (where the state of
(where the state of 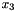 is missing).
is missing).
Expectation Maximization (.em) file format
This section describes the file format of .em files, which are used to specify a particular EM algorithm. The .em files are complementary to .fg files; in other words, an .em file without a corresponding .fg file is useless. Furthermore, one also needs a corresponding .tab file containing the data used for parameter learning.
An .em file starts with a line specifying the number of maximization steps, followed by an empty line. Then, each maximization step is described in a block, which should satisfy the format described in the next subsection.
Maximization Step block format
A maximization step block of an .em file starts with a single line describing the number of shared parameters blocks that will follow. Then, each shared parameters block follows, in the format described in the next subsection.
Shared parameters block format
A shared parameters block of an .em file starts with a single line consisting of the name of a ParameterEstimation subclass and its parameters in the format of a PropertySet. For example:
CondProbEstimation [target_dim=2,total_dim=4,pseudo_count=1]
The next line contains the number of factors that share their parameters. Then, each of these factors is specified on separate lines (possibly seperated by empty lines), where each line consists of several fields seperated by a space or a tab character. The first field contains the index of the factor in the factor graph. The following fields should contain the variable labels of the variables on which that factor depends, in a specific ordering. This ordering can be different from the canonical ordering of the variables used internally in libDAI (which would be sorted ascendingly according to the variable labels). The ordering of the variables specifies the implicit ordering of the shared parameters: when iterating over all shared parameters, the corresponding index of the first variable changes fastest (in the inner loop), and the corresponding index of the last variable changes slowest (in the outer loop). By choosing the right ordering, it is possible to let different factors (depending on different variables) share parameters in parameter learning using EM. This convention is similar to the convention used in factor blocks in a factor graph .fg file (see Factor block format).
Aliases file format
An aliases file is basically a list of "macros" and the strings that they should be substituted with.
Each line of the aliases file can be either empty, contain a comment (if the first character is a '#') or contain an alias. In the latter case, the line should contain a colon; the part before the colon contains the name of the alias, the part after the colon the string that it should be substituted with. Any whitespace before and after the colon is ignored.
For example, the following line would define the alias BP_SEQFIX as a shorthand for "BP[updates=SEQFIX,tol=1e-9,maxiter=10000,logdomain=0]":
BP_SEQFIX: BP[updates=SEQFIX,tol=1e-9,maxiter=10000,logdomain=0]
Aliases files can be used to store default options for algorithms.
 1.8.9.1
1.8.9.1